Recently, Professor Zhang Xiaofei's research group at the School of Mathematics and Statistics has made significant strides in the field of spatial transcriptomics. Their groundbreaking research paper titled"ENGEP: Advancing Spatial Transcriptomics with Accurate Unmeasured Gene Expression Prediction" was published in the esteemed journal Genome Biology (https://doi.org/10.1186/s13059-023-03139-w).
The study rigorously evaluated the efficacy of ENGEP using three distinct spatial transcriptomics datasets. The findings underscored ENGEP's remarkable performance, surpassing other methodologies by accurately predicting expression patterns of unmeasured genes in spatial context. This breakthrough holds immense promise in refining spatial transcriptomics data analysis.
Moreover, the research unveiled novel spatial expression patterns among the predicted unmeasured genes, shedding lights on the intricate functional architecture of complex tissues, such as the brain and tumors.
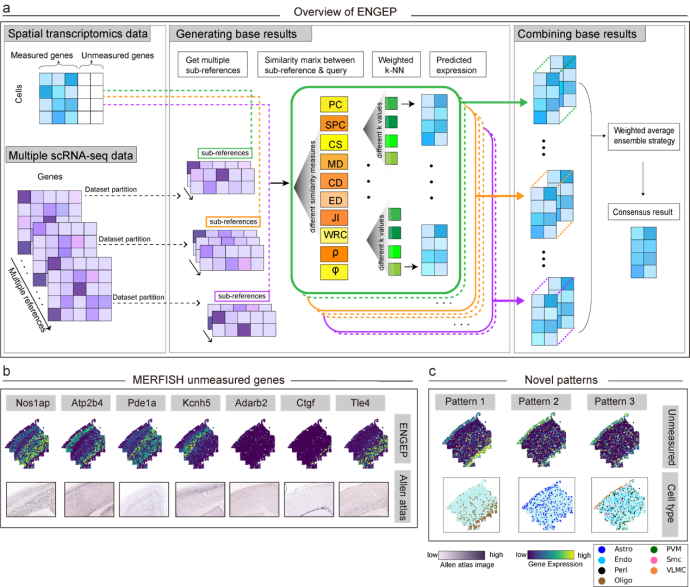
Overall, ENGEP adeptly addresses the challenge of numerous unmeasured genes in spatial transcriptomics data. Compared to alternative methods, ENGEP also excels in terms of runtime and memory efficiency, enabling seamless analysis of large-scale datasets.




